Our PCR-free Technology
A standard genotyping workflow. Single nucleotide specificity.
Gene editing validation redefined.
The Science Behind GeneAbacus
GeneAbacus empowers the gene editing community with speed, accuracy, and ease of use. Our core technology is based on PCR-free molecular tools developed over 20+ years of academic research to achieve precise and unbiased detection of DNA sequences. But how does it work? Let’s delve into the science behind GeneAbacus.
Padlock Probes: Highly Specific Detection of Gene Sequences
- Padlock probes are molecular locks that recognize and bind only to the intended sequences in your DNA samples.
- Each probe is custom-designed to perfectly match the unedited and/or the expected edited genotypes.
- Only when a padlock probe encounters its perfect match sequence, a high-fidelity enzyme called ligase, joins the two ends of the probe, forming a circular DNA molecule.
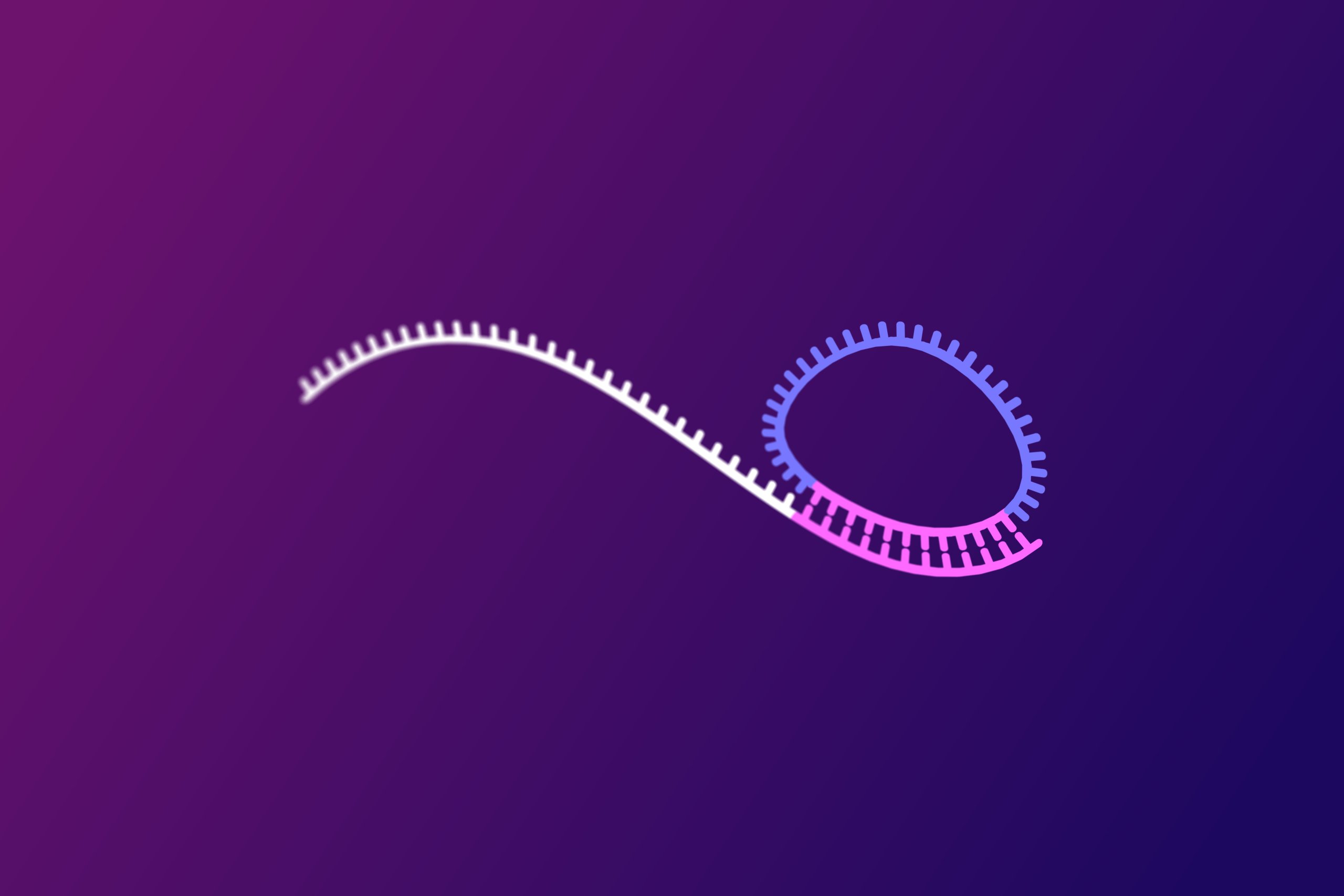

Rolling Circle Amplification: PCR-free DNA amplification
- Circularized padlocks are amplified by rolling circle amplification (RCA). A process that generates thousands of connected copies of the circularized probes.
- Resulting RCA products are individual sub-micron sized DNA molecules. Their large molecular size, facilitates downstream detection by low magnification fluorescence microscopy.
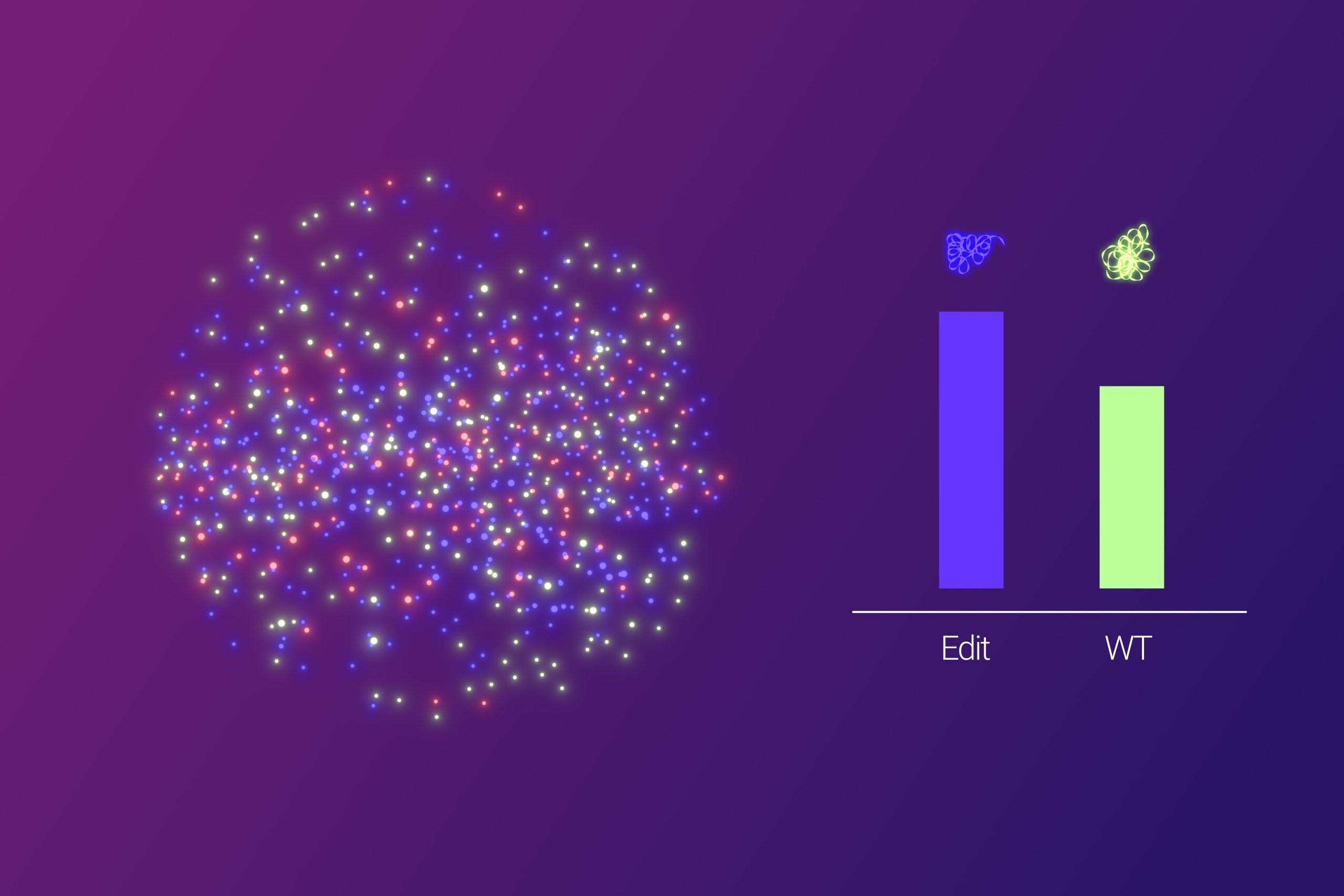
Amplified Single Molecule Quantification
- After RCA, every single amplified molecule is the result from one original gene sequence. This enables single molecule quantification analysis without the need for droplet compartmentalization, such as in digital PCR.
- Fluorescently-tagged oligonucleotides are used to differentiate RCA products stemming from the original wild type and edited sequences.
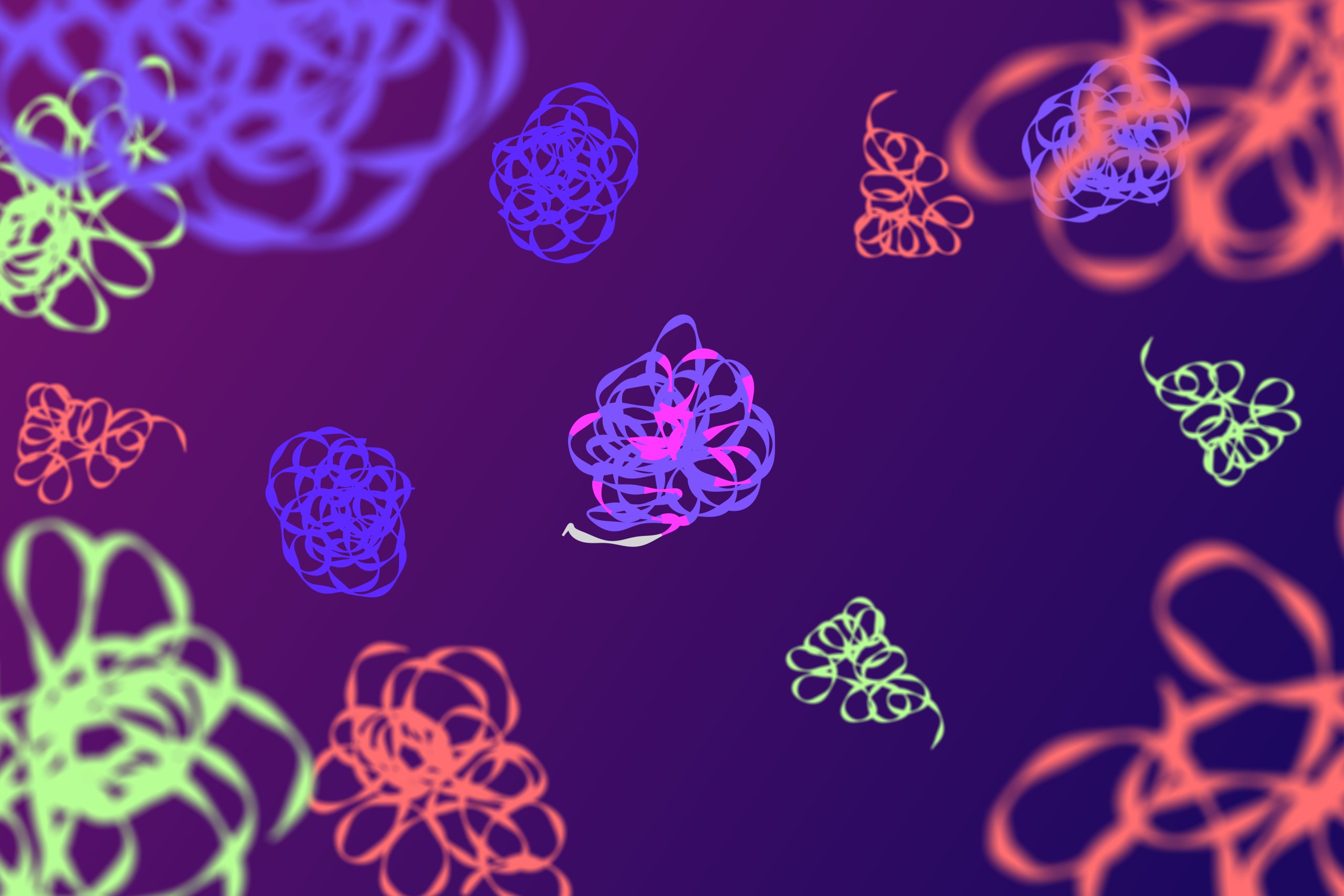
Highly Efficient Readout and Analysis
- Fluorescently barcoded RCA products are pulled and captured into an area equal to a single field of view of a low magnification microscope objective.
- This enables highly efficient imaging and fast computational image analysis.
- Our companion software, GAIA, processes resulting microscopy images, quantifies fluorescent signals and provides gene editing efficiencies in a user-friendly interface.
GeneAbacus: Accelerating Gene Editing Breakthroughs
By combining padlock probes, rolling circle amplification, and our unique single molecule readout technology, GeneAbacus offers a novel genotyping approach to gene editing validation. The GeneAbacus kit, is a bundle of reagents, consumables and software. This streamlined solution empowers gene editing scientists with the speed, accuracy, and ease of use needed to accelerate gene editing validation workflows.

