Our Technology
PCR-free assay. Single nucleotide precision. Gene editing analysis redefined.
Unveiling the Science Behind GeneAbacus
Gene editing holds immense promise for revolutionizing healthcare, but ensuring its effectiveness hinges on precise analysis of the edits made. Countagen’s GeneAbacus technology empowers researchers with a groundbreaking solution that delivers speed, accuracy, and ease of use. Countagen’s technology is based on molecular tools developed over 20+ years of academic research to achieve precise and unbiased detection of DNA sequences. But how does it work? Let’s delve into the science behind GeneAbacus.
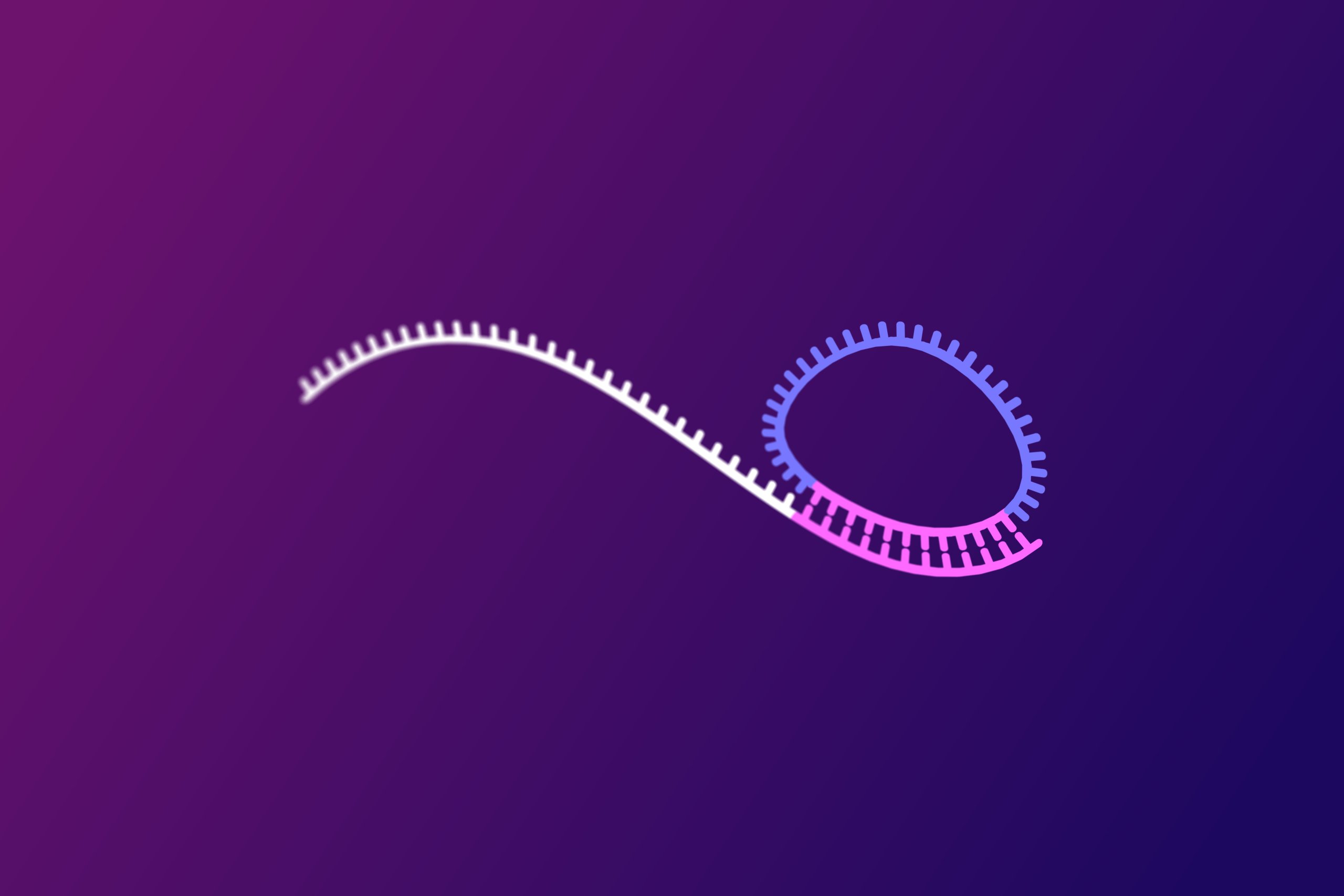
Padlock Probes: Targeting the Edited Sequence
- Imagine tiny molecular locks specifically designed to recognize and bind only to the intended edited sequence in your DNA. These are the padlock probes at the heart of GeneAbacus.
- Each probe is a single-stranded oligonucleotide, a short piece of DNA, custom-designed to perfectly match either the wild type (unedited) sequence or the edited DNA sequence you’re targeting.
- When a padlock probe encounters its perfect match in the genomic DNA, a high-fidelity enzyme called a ligase joins the two ends of the probe, forming a circular DNA molecule.

Rolling Circle Amplification: Linear Signal Generation
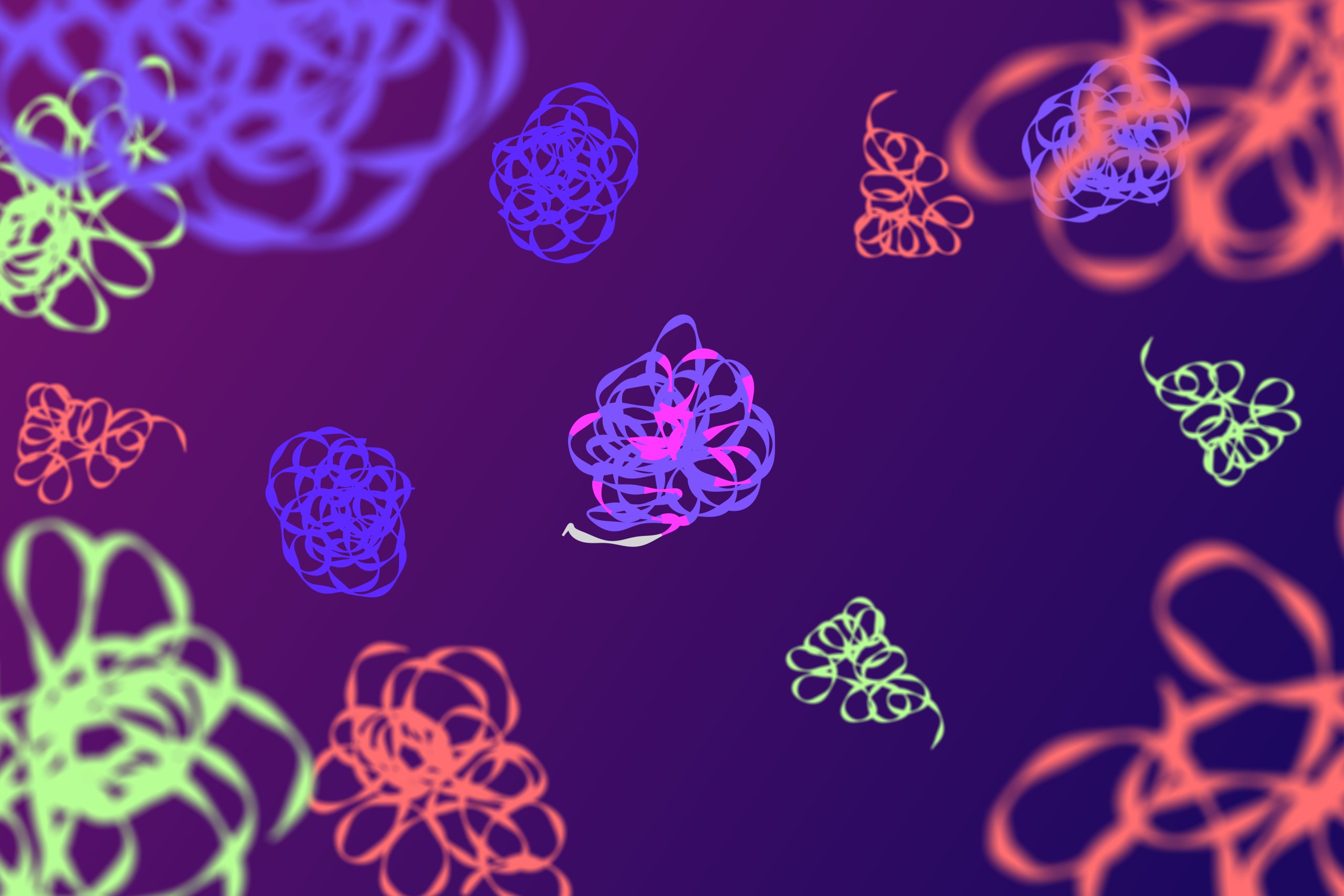
- Once the padlock probe is circularized, it undergoes a process called rolling circle amplification (RCA). This technique amplifies the signal from the bound and reacted probe, allowing for easy detection.
- A specialized highly processive DNA polymerase continuously synthesizes a long, single-stranded DNA molecule using the circular probe as a template. This process generates thousands of copies of the probe sequence.
- These individual concatenate DNA molecules collapse into sub-micron sized DNA balls, also called RCA product, that can include specific detection tag sequences that can be used to create a detectable signal.
Amplified Single Molecule Quantification with Single-Nucleotide Specificity: Ensuring Accuracy
- The power of GeneAbacus lies in its unmatched single-nucleotide specificity. Any mismatch between the probe and the DNA sequence, even a single nucleotide difference, prevents the ligase from joining the probe ends.
- This ensures that only perfectly matched sequences undergo RCA, resulting in a highly accurate representation of the editing efficiency and specificity.
- Each generated RCA product stems from one target sequence of interest enabling accurate digital nucleic acid quantification without the need for compartmentalisation, such as in digital PCR.
Streamlined Workflow and Analysis
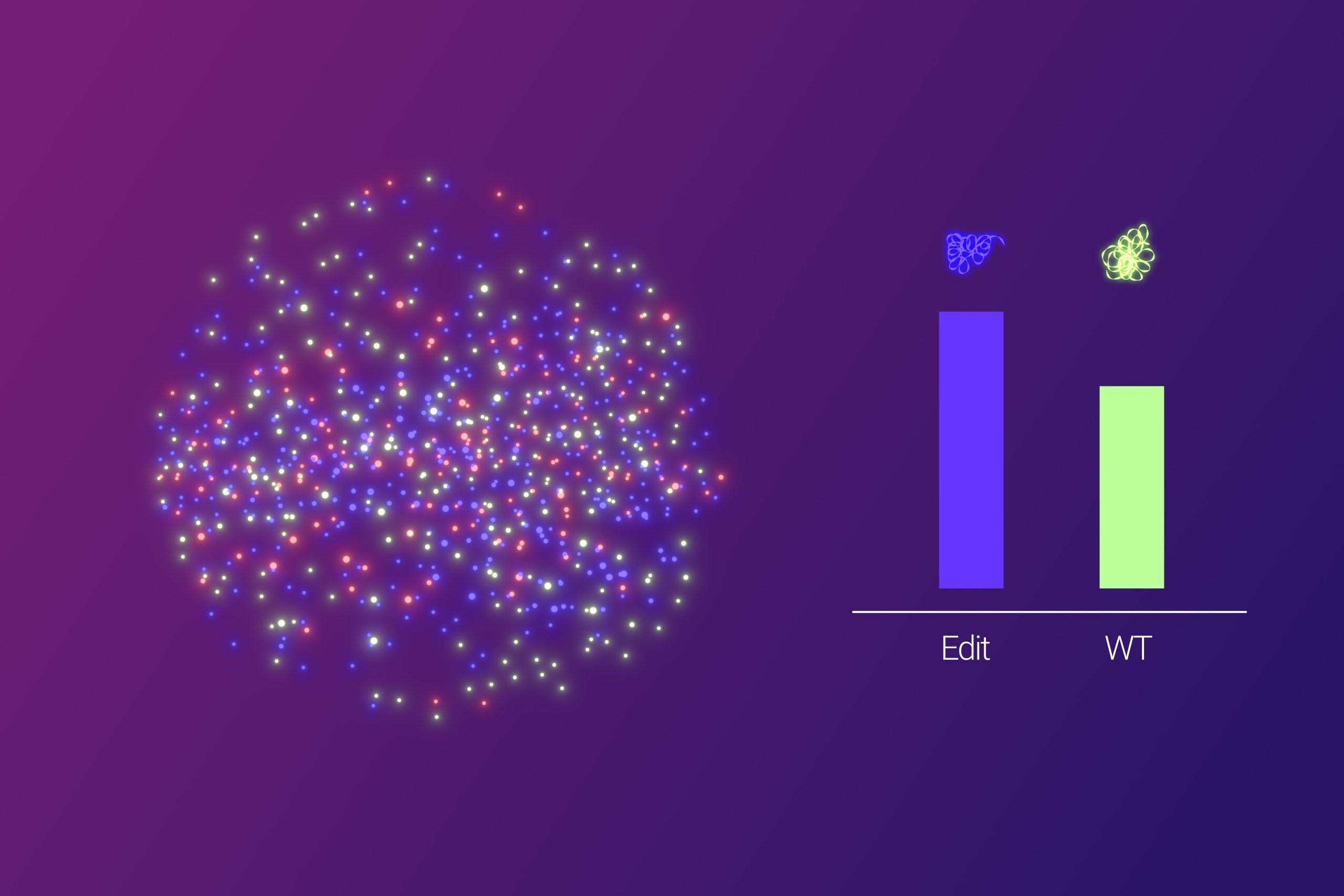
- Following amplification, fluorescent dyes are used to differentiate between probes targeting the wild type sequence, the edited sequence(s), and an internal control.
-
Labeled molecules are pulled and captured into the area of a single field of view of a low magnification microscope objective. Enabling highly efficient imaging acquisition and fast computational image analysis.
-
A dedicated image analysis software (GAIA, short for GeneAbacus Image Analyzer) then automatically quantifies the fluorescent signals, providing researchers with a clear picture of editing efficiency.
GeneAbacus: A Powerful Tool for Advancing Gene Editing Research
By combining padlock probes, rolling circle amplification, and Countagen’s unique enrichment technology, GeneAbacus offers a novel approach to gene editing analysis. GeneAbacus, is a bundle of reagents, consumables and software for gene editing efficiency quantification. This innovative technology empowers researchers with the speed, accuracy, and ease of use needed to accelerate breakthroughs in gene editing therapies.

