Content Overview
- The Importance of Accurate Analysis and Validation in Gene Editing
- Understanding the Limitations of PCR
- Introducing RCA Technology: What is it?
- Key Advantages of RCA Technology
- Applications of RCA in Gene Editing Validation
- GeneAbacus: An RCA-based Solution for Gene Editing Validation
- Future Implications
Scientists have made remarkable strides in deciphering the code of life, a journey that has since revolutionized healthcare. With the emergence and advancements of gene editing technologies like CRISPR/Cas9, diseases can potentially be cured rather than just treated. While the field is moving fast, the development of those therapies still takes a long time. Especially the early R&D phase as it is an iterative process of Discovery and Preclinical validation. A pivotal requirement and critical first step for enabling responsible gene editing is the validation of the gene edit, meaning detecting on- and off-target mutations. Recognizing the importance of accurate analysis and validation, our first blog post describes the limitations of traditional detection methods, and introduces a revolutionary technology – Rolling Circle Amplification (RCA).
The Importance of Accurate Analysis and Validation in Gene Editing
Accurate analysis and validation are crucial in the field of gene editing. Scientists check for on- and off-target mutations in gene knock-out and knock-in experiments to ensure the accuracy and specificity of their CRISPR/Cas9 editing. Checking for on-target mutations confirms that the target gene is altered as intended. On the other hand, monitoring off-target mutations is crucial to maintain the integrity of the genome and prevent unintended alterations that could lead to undesired effects in the development of new therapies.
Understanding the Limitations of PCR
Today, the most commonly used methods for detecting on- and off-target mutations are all PCR-based. They tend to underestimate the frequency of on-target activity due to decreased sensitivity for large deletions and less efficient amplification of large insertions[1]. Another constraint is bias towards more abundant templates[1]. This can lead to an overrepresentation of high copy number templates, potentially masking the presence of low abundant sequences.
Another limitation inherent to PCR is the highly demanding yet critical PCR optimization step. The need for high sensitivity complicates the optimization of reaction conditions, particularly for difficult templates like GC-rich regions[2]. Various parameters, including DNA template concentration, primer design, and PCR thermal cycling conditions, affect the accuracy and efficacy of amplification. As a consequence, ineffective amplification may result in false negatives or artifacts due to undesired secondary reactions. This can be particularly problematic in gene editing experiments where precise analysis and validation are essential.
In contrast, Rolling Circle Amplification (RCA) technology is PCR-free. This highly specific alternative to PCR is thus able to overcome the limitations of PCR while improving precision. At Countagen, our research has demonstrated that our RCA-based method yields comparable editing efficiencies to established gold standard PCR-based methods.
Introducing RCA Technology: What is it?
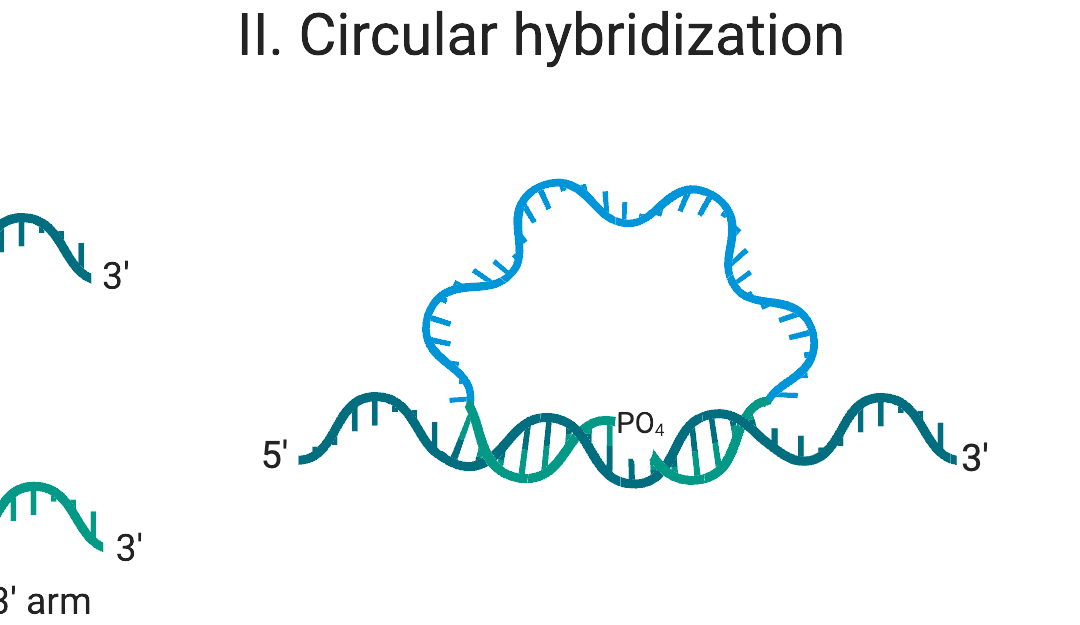
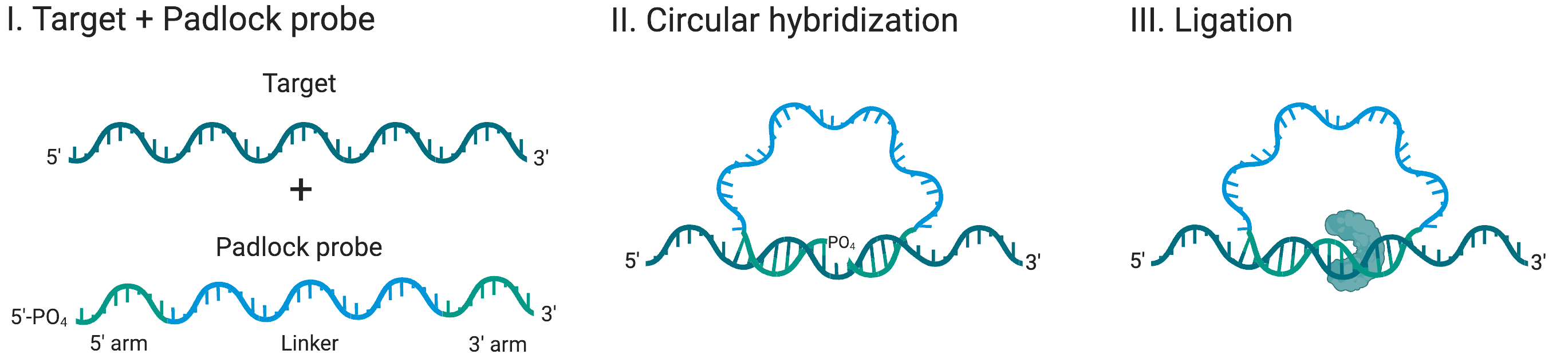
RCA is an isothermal nucleic acid amplification technique that permits single-molecule counting[3]. In RCA, a circular template of DNA is replicated as a long single-stranded DNA concatemer, spooling off during amplification (Figure 1). The result is DNA nano-balls with single-molecule integrity – making RCA ideal for digital nucleic acid quantification. Due to its isothermal nature, RCA occurs at one temperature, eliminating the need to cycle or have very accurate temperature control like PCR.
Figure 1: Padlock probe binding to the single-stranded region of the target DNA. Created with BioRender.com.
RCA often uses padlock probes[4], as seen in Figure 1, which are linear oligonucleotides with a 5’ phosphate group, designed to have complementary regions to a given target at their 5’ and 3’ ends. Upon hybridization to the target, the probe forms a circular structure with a nick between the ends. Only in the event of a perfect sequence match, a high-fidelity ligase joins the ends of the probe, “locking” the probe onto its target. This powerful combination enables differentiation of single base mismatches. Subsequently, the circular probe is used as a template for DNA synthesis by a high processivity DNA polymerase.
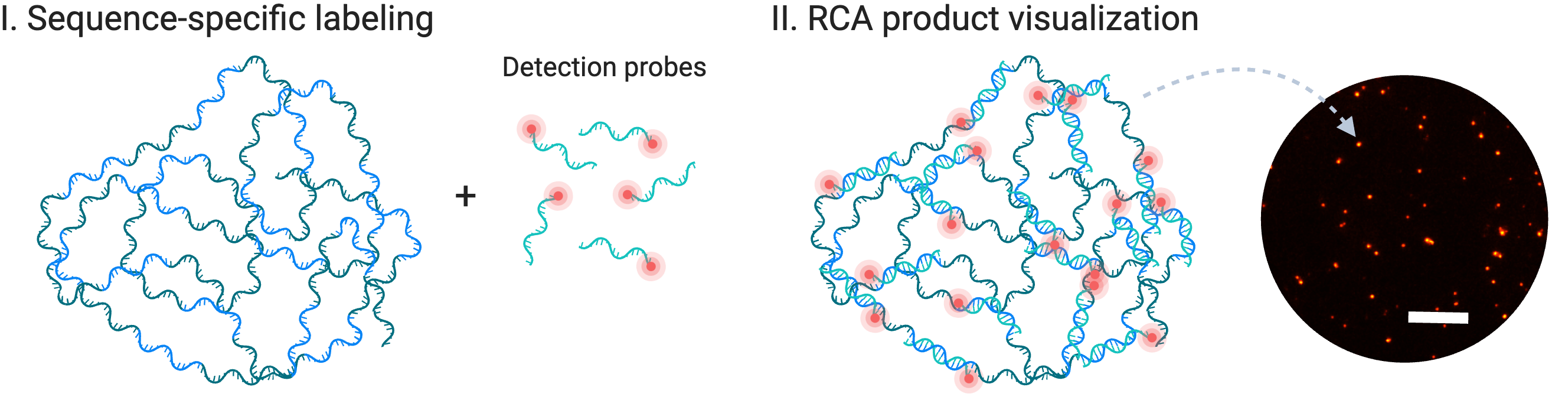
Figure 2: Visualization of a single RCA product that is fluorescently labeled, allowing for fluorescence detection. Created with BioRender.com.
Another added benefit of padlock probes is that the linker between their 5’ and 3’ arms can be designed freely to offer a range of possibilities for detecting RCA products[5]. Some examples include fluorescence (Figure 2) and colorimetric detection. Besides that, padlock probes provide a unique advantage by offering sequence identity independence. This means that even sequences with extreme GC content, often challenging for PCR amplification, can be processed easily.
Key Advantages of RCA Technology
RCA technology offers several key advantages for gene editing analysis and validation:
- Ensures single nucleotide specificity: Padlock probes confer single nucleotide specificity, ensuring that only the desired DNA sequence is amplified, minimizing the risk of false-positives.
- Eliminates need for specific primers: Unlike PCR, RCA does not require specific primers for initiating amplification, eliminating concerns associated with primer design issues and target sequence variations, which enhances overall reliability.
- Eliminates amplification bias: The sequence context independence of padlock probes eliminates the risk of amplification bias or cross-contamination in PCR.
- Enables single molecule detection: Single molecule quantification assays are known to provide the ultimate sensitivity and precision for molecular analysis[6]. RCA’s single molecule detection reinforces this quality, strengthening the credibility of RCA-based methods in accurately counting gene editing events. This allows scientists using RCA-based methods to confidently screen their pools and move on to functional study. Additionally, since single molecule detection (like RCA) is the highest resolution measurement one can make [7], it is beneficial in quantifying gene editing events, or studying heterogeneity within a sample.
- Saves time and costs: RCA technology simplifies the validation workflow by eliminating the need for primer design and optimization. Its isothermal nature removes the need for expensive thermocyclers or bioinformaticians.
With these advantages, RCA technology is redefining how gene edits are analyzed, providing scientists with a reliable and cost-efficient method.
Applications of RCA in Gene Editing Validation
RCA has diverse applications in gene editing validation such as the detection of gene mutations, verification of gene editing outcomes, quantification of gene editing efficiency, screening of gene editing tools, analysis of pooled clones, and digital nucleic acid quantification. In general, RCA technology can enhance the accuracy, specificity, and efficiency of gene editing validation, making it a cost-effective tool for scientists in the field of molecular biology and genetic engineering.
GeneAbacus: An RCA-based Solution for Gene Editing Validation
At Countagen, our RCA-based solution, GeneAbacus™, is a ready-to-use bundle of reagents, consumables and software for gene editing efficiency quantification. It reliably quantifies editing efficiency in cell pools as well as the subsequent analysis of clones or organisms within 5 hours, as compared to days or even weeks. This same-day solution accelerates the analysis process, and its flexibility enables the analysis of up to 32 samples per day under the same condition, regardless of the origin and sequence characteristics. The analysis outcome is generated by counting the individual number of fluorescently-labeled amplicons through our automated image analysis software. Thus, the results provide higher accuracy than analog measurements like measuring intensities of a gel electrophoresis band or quantitative PCR reaction.
Future Implications
The application of RCA technology in gene editing validation opens up new possibilities for scientific research and medical advancements. As scientists continue to explore the potential of gene editing for various applications, the need for accurate analysis and validation becomes increasingly important. With its highly specific amplification, RCA offers a reliable and efficient solution for addressing this need. It paves the way for quicker and more precise gene editing workflows, ultimately aiding advancements in fields such as personalized medicine, agriculture, and synthetic biology.
In conclusion, our RCA-based kit, GeneAbacus, is redefining how validation is carried out in gene editing experiments. By overcoming the limitations of PCR, this highly specific alternative accurately quantifies editing efficiency (in cell pools, cell clones, or even organisms), providing actionable percentage efficiencies for informed cell or model organism selection in a cost-efficient manner.
Accelerate your gene editing research by ordering GeneAbacus today.
An RCA-based Solution: GeneAbacus
Redefine how you validate your gene edits

References
[1] Zischewski J, Fischer R, Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnology Advances [Internet]. 2017 Jan [cited 2024 Feb 9];35(1):95–104. Available from: https://www.sciencedirect.com/science/article/pii/S0734975016301586 doi:10.1016/j.biotechadv.2016.12.003
[2] Obradovic J, Jurisic V, Tosic N, Mrdjanovic J, Perin B, Pavlovic S, et al. Optimization of PCR conditions for amplification of GC-richegfrpromoter sequence. Journal of Clinical Laboratory Analysis [Internet]. 2013 Nov [cited 2024 Feb 9];27(6):487–93. Available from: https://pubmed.ncbi.nlm.nih.gov/24218132/ doi:10.1002/jcla.21632
[3] 2. Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genetics [Internet]. 1998 Jul [cited 2024 Feb 9];19(3):225–32. Available from: https://www.nature.com/articles/ng0798_225#citeas doi:10.1038/898
[4] Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. Padlock Probes: Circularizing Oligonucleotides for Localized DNA Detection. Science [Internet]. 1994 Sept 30 [cited 2024 Feb 9];265(5181):2085–8. Available from: https://www.science.org/doi/10.1126/science.7522346 doi:10.1126/science.7522346
[5] Neumann F. Advancing isothermal nucleic acid amplification tests : Towards democratization of diagnostics [Internet] [PhD dissertation]. [Stockholm]: Department of Biochemistry and Biophysics, Stockholm University; 2020. Available from: https://urn.kb.se/resolve?urn=urn:nbn:se:su:diva-185978
[6] Kühnemund M, Hernández-Neuta I, Sharif MI, Cornaglia M, Gijs MAM, Nilsson M. Sensitive and inexpensive digital DNA analysis by microfluidic enrichment of rolling circle amplified single-molecules. Nucleic Acids Research [Internet]. 2017 Jan 10 [cited 2024 Feb 9]; Available from: https://academic.oup.com/nar/article/45/8/e59/2888443 doi:10.1093/nar/gkw1324
[7] Walt DR. Optical methods for single molecule detection and analysis. Analytical Chemistry [Internet]. 2012 Dec 10 [cited 2024 Feb 9];85(3):1258–63. Available from: https://pubs.acs.org/doi/10.1021/ac3027178 doi:10.1021/ac3027178
Newsletter
Subscribe now so you won’t miss out on the newest and most exciting content!